The minimum input to dafmpb is the PQR file.
DAFMPB can read meshes generated from MSMS or TMSMesh. If no input mesh is provided, DAFMPB will invoke the built-in surface meshing routine.
- Example 1, use built-in mesh routine
> ./dafmpb --pqr-file=GLY.pqr - Example 2, use mesh generated by MSMS
> ./dafmpb --pqr-file=GLY.pqr --mesh-format=1 --mesh-file=GLY.pqr-mesh.data-d20-r0.5 - Example 3, use mesh generated by TMSmesh
> ./dafmpb --pqr-file=fas2.pqr --mesh-format=2 --mesh-file=fas2.off
A list of command line options to the program can be found by issuing
> ./dafmpb --help
To launch dafmpb on a cluster with Slurm workload manager, a job script looks like this
#! /bin/bash -l
#SBATCH -p queue
#SBATCH -N 2
#SBATCH -t 00:10:00
srun -n 2 -c 48 ./dafmpb --pqr-file=fas2.pqr.ext --mesh-format=2 --mesh-file=fas2.off --hpx-threads=24
The -c option equals to the number of cores Slurm sees on each compute node and the --hpx-threads
option equals the number of physical cores available on the compute node.
If the above cluster were using the PBS workload manager, the script looks like this
#! /bin/bash -l
#PBS -l nodes=2:ppn=48
aprun -n 2 -d 48 ./dafmpb ...
Finally, the --potential-file option allows user to specify an output stream to hold the
results of the potentials. The output can be then visualized using VCMM.
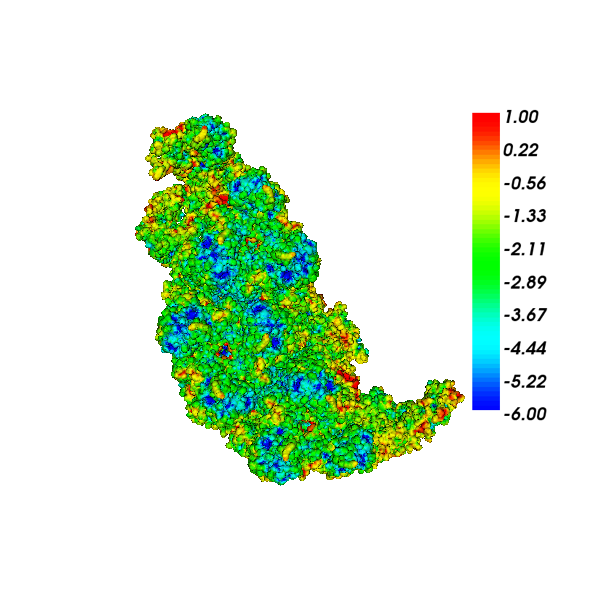
This picture shows the surface potential of the aquareovirus virion, 3K1Q, consisting of 203,111 atoms. The surface mesh was generated using TMSMesh, consisting of 7,788,246 triangle elements and 3,888,281 nodes. The results were computed requiring 6-digit accuracy on DASHMM. The polar energy is -8.81808e4 kcal/mol and the total solvation energy is -2.42747e4 kcal/mol.